A Random Mixture Model for Graphs
Working on a Mixture Model for the edges of a graph
A Mixture Model for Random Graphs
Abstract
Traditional graph analysis methods predominantly focus on node degree to infer graph structure, typically grouping nodes into classes based on high interaction rates within each class. However, this approach often fail to capture the underlying structure of a graph. In this report, we delve into a mixture model that shifts the focus from nodes to edges, thereby offering a more nuanced depiction of graph structure. Our study emphasizes the importance of parameter initialization in the algorithm and demonstrates its application through the analysis of two real-world datasets. We explore how this edge-centric approach provides a detailed understanding of graph topology, capturing complex structures that node-based methods may miss. However, the effectiveness of this method is somewhat constrained by the graph’s size. This limitation highlights the need for further refinement to extend its applicability to larger networks.
The repository is the one we produced for our project on the paper : Franck Picard Daudin, J-J. and Stéphane Robin. (2008). A mixture model for random graphs.
We did this project in the MVA course “Introduction to Probabilistic Graphical Models and Deep Generative Models”, teached by P. Latouche, and P.A. Mattei, and with R. Khellaf as Teaching Assistant. You can read the report we made of this project here
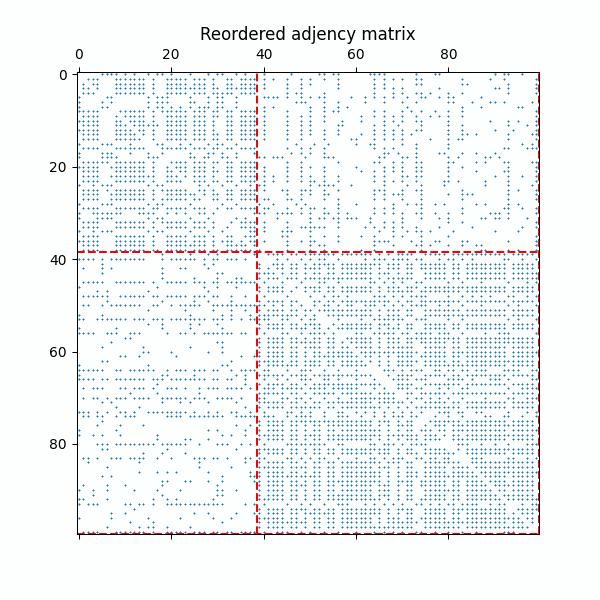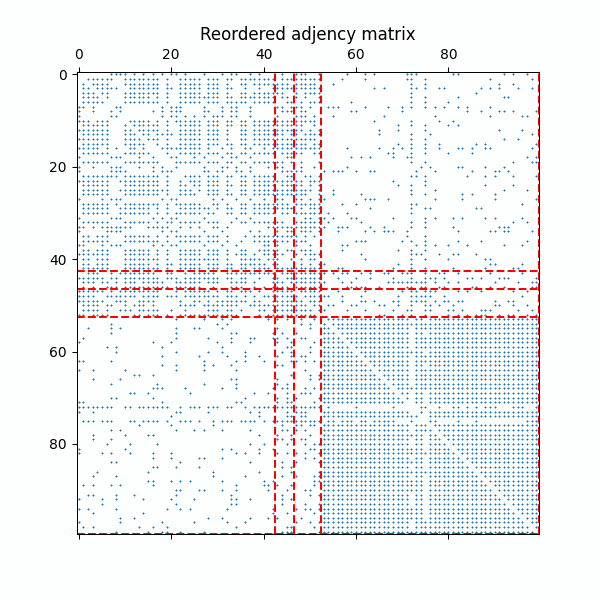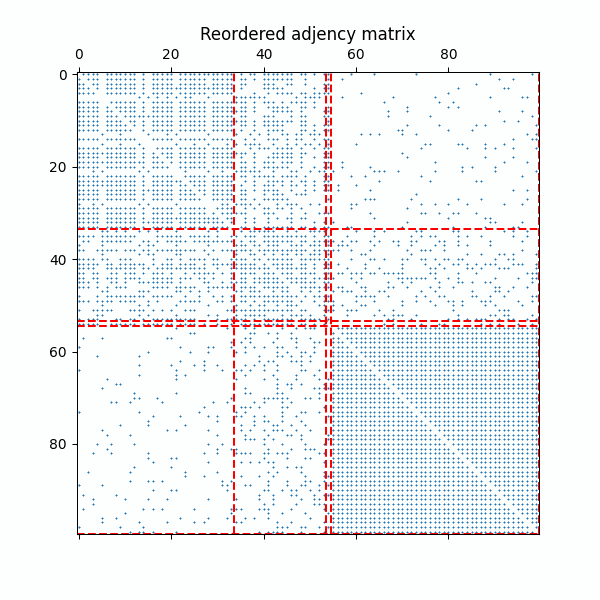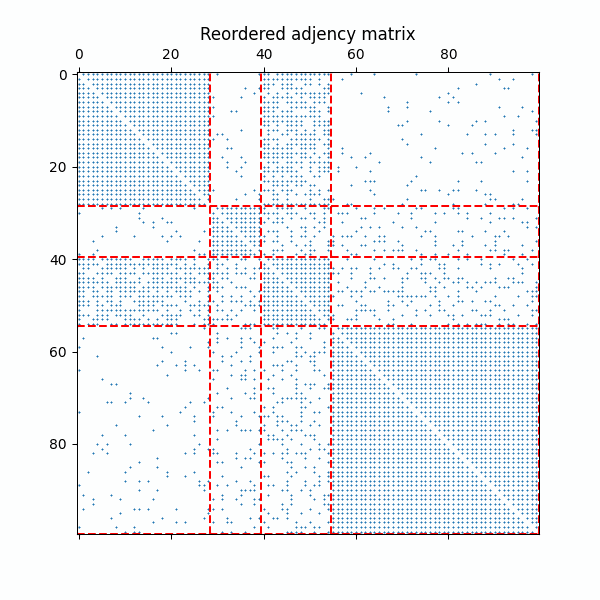
Example of the reordered adjency matrix with as the ERMG model converges
Project of Alexandre François and Sacha Braun
Installation
git clone https://github.com/ElSacho/Mixture4graph
cd Mixture4graph
pip install -r requirements.txt
General structure
We choose to implement the EM algorithm using torch tensor, because it allows to leverage GPU capacities and to fasten the computation time. Everything is implement from scratch, using torch, or numpy. We also propose a numpy version of the EM algorithm in the file old/em.py. We also tried to produce a docstring on most of the used methods of the EM_torch.py file, if you need further information on a precise method.
How it Works - The notebook main_notebook.ipynb
You can find most of the following explanations in the notebook file.
-
Generate Graphs
If you want to generate a graph, you need to provide the model with the prior (\pi), (\alpha), and the number of vertices you want. You can call the generator with the functiongeneratethat will return the generated graph, orgenerate_and_give_tauthat will return a tuple with the generated graph, and a vector to explain to which clusters all nodes belong from the generator file. -
Estimate Parameters
- To estimate the parameters of a graph, you first need to initialize a model. To do this, you can call the
mixtureModelclass. \ - Then, you can fit the data with the model by calling the
fitfunction. You need to specify the number of clusters you want for your fit, and you can also provide the model with a list of number of clusters. You may want to add an initialization method for the first values of (\tau), to test the model with several initialization methods. - If you want to iterate the algorithme to stabilize the results, you can use the other function `precise_fit that will iterate the fit function for nbr_iter_per_cluster time per cluster, in order to reduce and show the variance of the different methods. As a parameter, you can give a list of method that you want to use, the string argument must be ‘modularity’, ‘spectral’,’random’, ou ‘hierarchical’. In our experiments, we only studied the first three of them as the results from ‘hierarchical’ were not really interesting.
- To estimate the parameters of a graph, you first need to initialize a model. To do this, you can call the
-
Plot the results
All the functions to plot the results are in the mixtureModel class.-
plotJRXwill plot the values of the (\mathcal{J}(R_{\mathcal{X}})) for all the clusters that has been studied -
plotICLwill plot the values of the ICL which is the value that we use to find the good number of clusters -
plot_adjency_matrixwill plot the adjency matrices of the final model. If you do not precise any value for the number of clusters, it will plot all the adjency matrix, or just one if you precise the one you want. You may also want to see the labels of the nodes in the adjency matrix, that you can do using using show_names = True.
-
-
Generate a new graph
- Finaly, you can generate a new graph with the values of the model. To access those values, you can use the dictionary of the mixtureModel, with the number of clusters you need. It will give you another dictionnary with the returned ‘tau’, ‘pi’, ‘prior’ .. values that you can use.
- You can finish ploting the orignal graph and the generated one. To do so, use the function show_colors_multiple_graph, were you give the two graphs, their respective tau values (to add the color on the clusters) and the titles that you want for your two graphs.
How it Works - Running the code on ssh
You can also run one function of the code that will produce all the results presented in the report. To do so, you can use the file `ssh_initialisation.py. When running this file, you will need to write the name of the output folder in which you want your figures to be saved, and the data that you want to use. A simple reading of the code allows to understand how you can use the created code for your own data : just read your data with nx.
VisualizeData
We also created a class VisualizeData that you can use as VisualizeData.print_generalities(graph) to print the most interesting information of the graph, or VisualizeData.plot_figure_graph(graph) to plot the degree distribution of the graph.
DataSets
You can find several datasets in the data folder. If we created the dataset from scratch, the functions we used to create this dataset are provided in the corresponding folder. The main datasets we used are:
-
The coappearance network of characters in the novel Les Misérables by Victor Hugo. This dataset includes 77 nodes representing characters from Les Misérables and 254 edges. Each edge connects two characters that share a common scene in the book.
-
An X (formerly Twitter) interaction network for the 117th United States Congress House of Representatives. This was constructed using members’ official Twitter handles. The Twitter API was employed to obtain all Tweets by members of Congress from February 9, 2022, to June 9, 2022. Each node represents the account of a member of Congress, connected to others through interactions such as replies, retweets, or comments on each other’s tweets. The network consists of 475 nodes and 10,222 edges.
Next step
Some next contributions of the project could include the implementation of new clustering initialization method, in the file initialisation_methods.py , or other criterion to choose the number of clusters such as BIC, AIC or anything else.